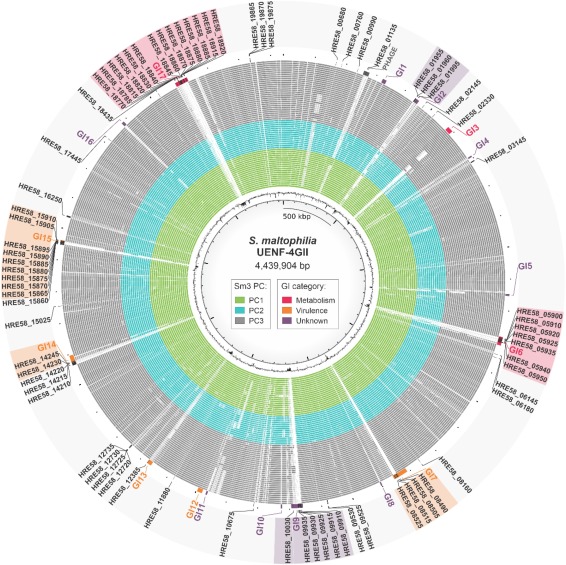
The Stenotrophomonas maltophilia complex (Smc) is a cosmopolitan bacterial group that has been proposed an emergent multidrug-resistant pathogen. Taxonomic studies support the genomic heterogeneity of Smc, which comprises genogroups exhibiting a range of phenotypically distinct strains from different sources. Here, we report the genome sequencing and in-depth analysis of S. maltophilia UENF-4GII, isolated from vermicompost. This genome harbors a unique region encoding a penicillin-binding protein (pbpX) that was carried by a transposon, as well as horizontally-transferred genomic islands involved in anti-phage defense via DNA modification, and pili glycosylation. We also analyzed all available Smc genomes to investigate genes associated with resistance and virulence, niche occupation, and population structure. S. maltophilia UENF-4GII belongs to genogroup 3 (Sm3), which comprises three phylogenetic clusters (PC). Pan-GWAS analysis uncovered 471 environment-associated and 791 PC-associated genes, including antimicrobial resistance (e.g. blaL1 and blaR1) and virulence determinants (e.g. treS and katG) that provide insights on the resistance and virulence potential of Sm3 strains. Together, the results presented here provide the grounds for more detailed clinical and ecological investigations of S. maltophilia.
Link of publication HERE
Publications 15
Research 2
 Posted on 08 Dec 2021
Posted on 08 Dec 2021
 Posted on 03 Nov 2021
Posted on 03 Nov 2021
New paper published! - Phylogenetic analysis and population structure of Pseudomonas alloputida
Posted on 03 Nov 2021 Posted on 03 Nov 2021
Posted on 03 Nov 2021