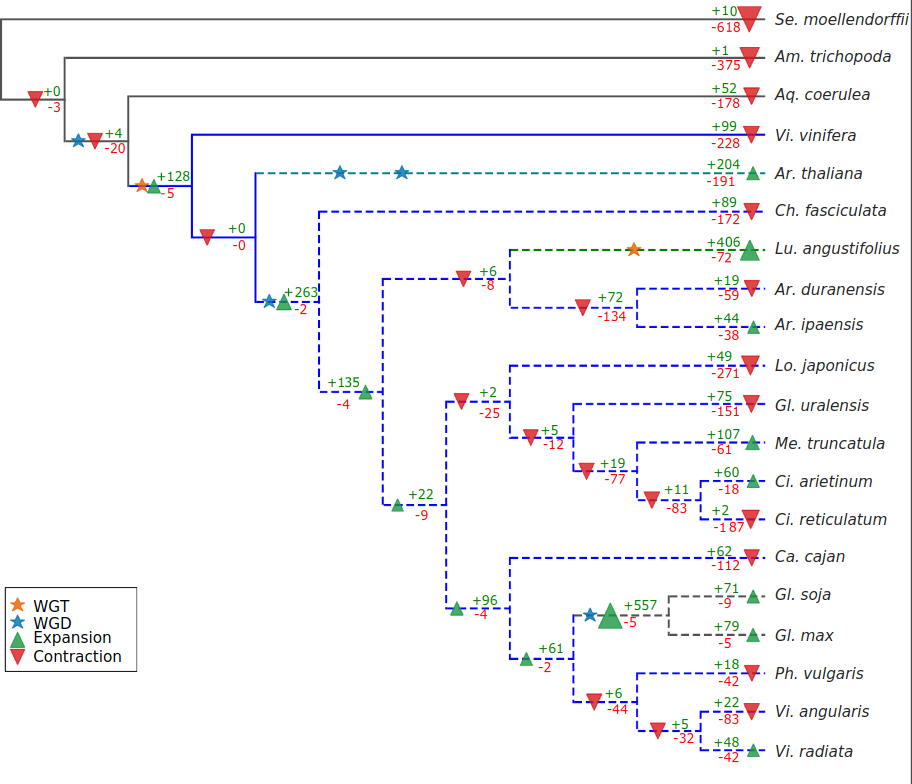
Transcription factors (TF) are essential for plant growth and development. Several legumes (e.g. soybean) are rich sources of protein and oil, with great economic relevance. Here we report a phylogenomic analysis of TF families in legumes and their potential association with important traits (e.g. nitrogen fixation). We used TF DNA‐binding domains to systematically screen the genomes of 15 legume and 5 non‐legume species. TF orthologous groups (OG) were used to estimate OG sizes in ancestor nodes using a gene birth‐death model, which allowed the identification of lineage‐specific expansions. OG analysis and rate of synonymous substitutions show that major TF expansions are strongly associated with whole‐genome duplication (WGD) events in the legume (~58 mya) and Glycine (~13 mya) lineages, which account for a large fraction of the Phaseolus vulgaris and Gl. max TF repertoires. Out of the 3407 Gl. max TFs, 1808 and 676 can be have homeologs within single syntenic regions in Ph. vulgaris and Vitis vinifera, respectively. We found a trend for TFs expanded in legumes to be preferentially transcribed in roots and nodules, supporting their recruitment early in the evolution of nodulation in the legume clade. Some families also showed count differences between G. max and the wild soybean Gl. soja, including genes located within important quantitative trait loci. Our findings strongly support the roles of two WGDs in shaping the TF repertoires in the legume and Glycine lineages, which are likely related to important aspects of legume and soybean biology.
Link of publication HERE
Publications 15
Research 2
 Posted on 08 Dec 2021
Posted on 08 Dec 2021
 Posted on 03 Nov 2021
Posted on 03 Nov 2021
New paper published! - Phylogenetic analysis and population structure of Pseudomonas alloputida
Posted on 03 Nov 2021 Posted on 03 Nov 2021
Posted on 03 Nov 2021