Frequently asked questions
- What is Raw read count? After RNAseq read mapping (*.BAM files)to a given list of genomic features (*.gff), we can count the number of reads mapped (uniquely mapping) to each feature, specially at the gene level. Essentially, total read count associated with a gene (meta-feature) = the sum of reads associated with each of the exons (feature) that "belong" to that gene.
-
What is read normalization?
Reads Per Kilobase Million (RPKM), Fragments Per Kilobase Million (FPKM), and Transcripts Per Million (TPM) are the most common units reported to estimate gene expression based on RNA-seq data. Both units are calculated from the number of reads that mapped to each particular gene sequence and both units are calculated taking into account two important factors in RNA-seq:
a. The number of reads from a gene depends on its length. One expects more reads to be produced from longer genes.
b. The number of reads from a gene depends on the sequencing depth that is the total number of reads you sequenced. One expects more reads to be produced from the sample that has been sequenced to a greater depth. -
What is the software pipeline used in this atlas?
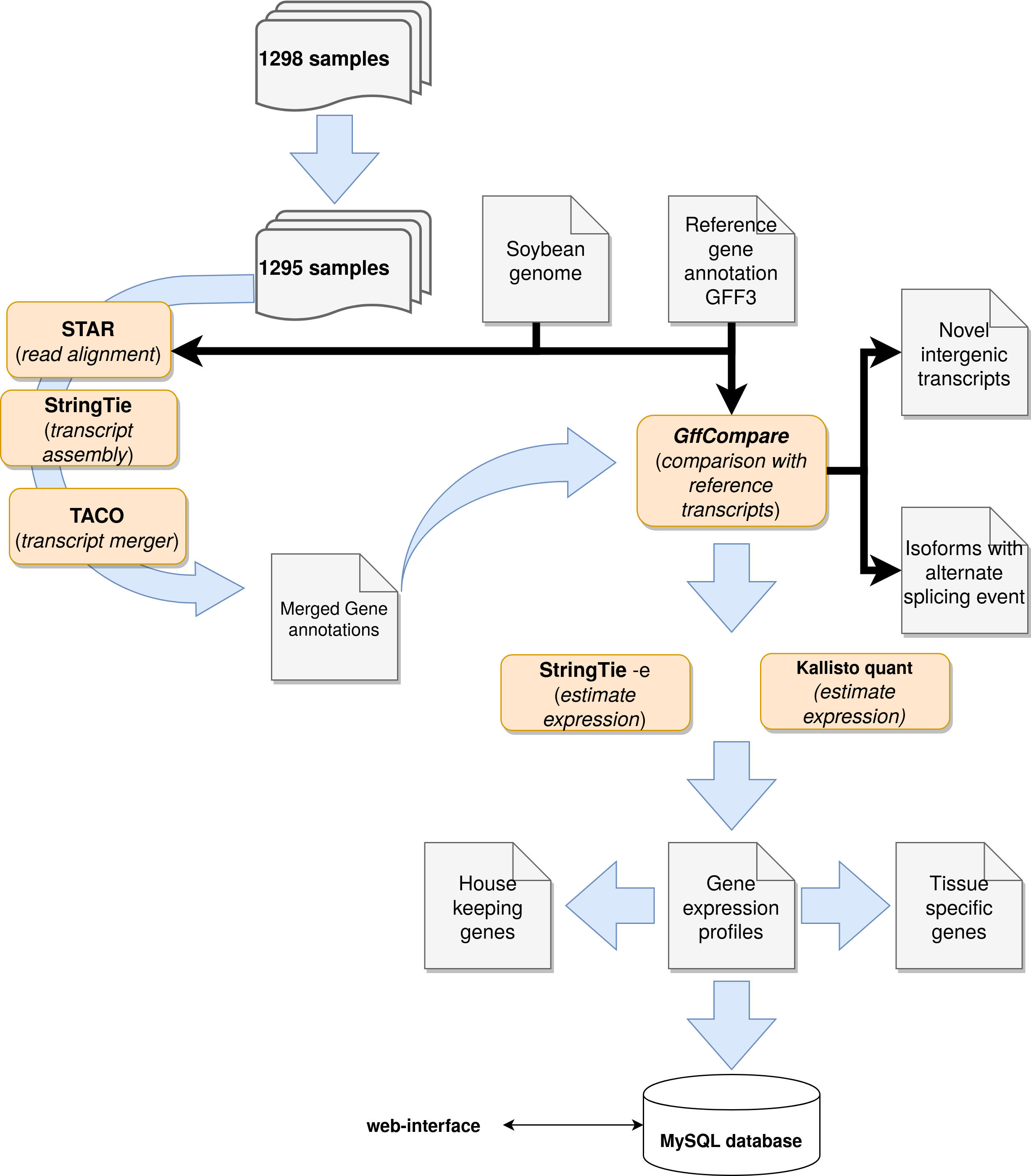
- Where can I find the scripts used in this pipeline? Soon we will make the scripts publicly available on Figshare.
- When I try to download datasets with (*.tsv) file extension, I see an error Try to use different web browser. Your anti-virus software might be blocking *.tsv file
- Internal Server Error!! Try to clear your browser cache,cookies and visit again
- Does this web portal use cookies? No. However, we use google analytics script to track unique number of visitors.
About Us
If you are using our dataset please cite us.
Systematic analysis of 1,298 RNA-Seq samples and construction of a comprehensive soybean (Glycine max) expression atlas.
Fabricio B. Machado, Kanhu Charan Moharana, Fabricio Almeida-Silva, Rajesh Kumar Gazara, Francisnei Pedrosa-Silva, Fernanda S. Coelho, Clicia Grativol, Thiago M. Venancio
bioRxiv 2019.12.23.886853; doi: https://doi.org/10.1101/2019.12.23.886853